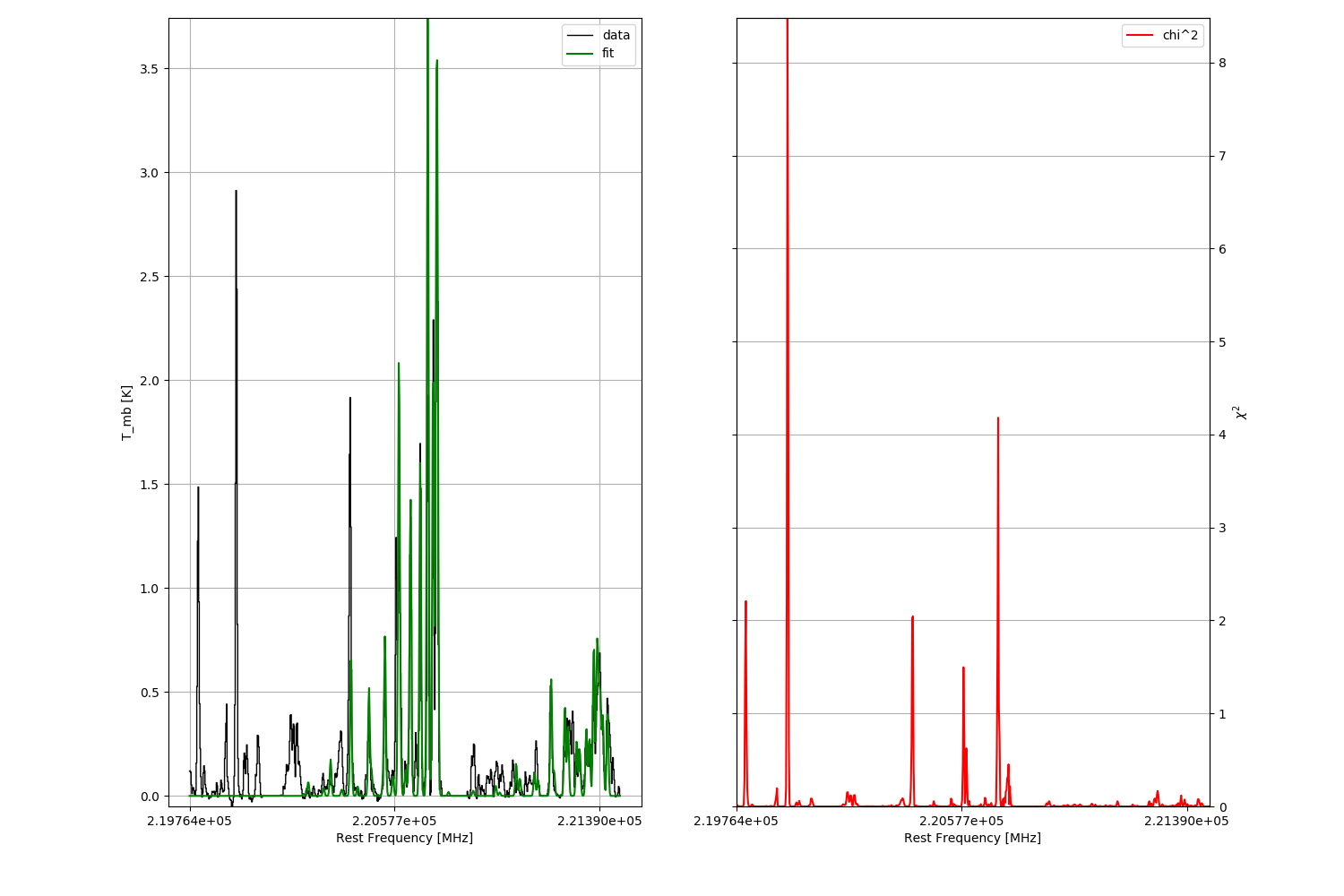
This function provides a simplified interface for MAGIX using the myXCLASS program. The function starts MAGIX using the Levenberg-Marquardt algorithm (with four processors) to fit experimental data with the myXCLASS program.
Input parameters:
-
NumberIteration: max. number of iterations (default: 50).
-
AlgorithmXMLFile: only necessary, if the user wants to use another fit algorithm (than Levenberg-Marquardt) for fitting. Therefore, the path and name of a MAGIX xml-file defining settings for an algorithm or algorithm chain has to be given.
-
MolfitsFileName: path and name of the extended molfit file, including the source size, the rotation temperature the column density, the velocity width, the velocity offset and the flag indicating if the current component is considered for core c or foreground f.In contrast to the format of the molfit file used for the myXCLASS function the extended molfit file required by the myXCLASSFit function contains one (three) additional column(s) for each parameter of each component. In order to fit the ratio(s) of isotopologues as well, the iso file has to include two additional columns
-
experimentalData: This parameter offers two different possibility to send the experimental data to the myXCLASSFit function:
-
the parameter experimentalData defines the path and name of and experimental xml-file suitable for MAGIX.
-
the parameter experimentalData defines the path and name of an ASCII file called observational data file, where the first column describe the frequency (in MHz) and the second column the beam temperature (intensity).
-
The following parameters are needed, if the parameter experimentalData} does NOT describe the path and name of a MAGIX xml-file:
-
vLSR: velocity (local standard of rest) in km s-1 (default: 0) used in the calculation of the synthetic spectra, see description for myXCLASS function. Please note, for the myXCLASSFit function XCLASS uses the user-defined lower and upper limits of a velocity offset parameter in the molfit file, if this parameter is fitted within a MAGIX run.) Please note, using a xml-file, the vLSR parameter can be defined for each obs. data file.
-
TelescopeSize: for single dish observations (Inter_Flag = F}): TelescopeSize describes the size of telescope (in m), (default: 1); for interferometric observations (Inter_Flag = T}): TelescopeSize describes the interferometric beam FWHM size (in arcsec), (default: 1).
-
Inter_Flag (T/F): defines, if single dish (F}) or interferometric observations (T}) are described, (default: F}).
-
t_back_flag (T/F): defines, if the user defined background temperature and temperature slope given by the input parameters tBack and tslope describe the continuum contribution completely (t_back_flag = T) or not (t_back_flag = F}) (default: T).
-
tBack: background temperature (in K), (default: 0).
-
tslope: temperature slope (dimensionless), (default: 0).
-
nH_flag: (T}/F}), defines, if column density, spectral index for dust and kappa are given by the molfit file (F) or, if nH_flag is set to T, the following three parameters define the H column density, spectral index for dust and kappa for all components (default: F):
-
N_H (has to be given only if nH_flag is set to T) Hydrogen column density (in cm-2), (default: 0).
-
beta_dust: (has to be given only if nH_flag is set to T) spectral index for dust (dimensionless), (default: 0).
-
kappa_1300: (has to be given only if nH_flag is set to T) kappa (cm2 g-1), (default: 0.01).
-
iso_flag: use isotopologues (T/F). If iso_flag is set to T} isotopologues defined in the iso ratio file are used (default: F).
-
IsoTableFileName (has to be given only if iso_flag is set to T): path and name of an ASCII file including the iso ratios between certain molecules. If no file name is given (default), the so-called iso-flag is set to F.
The following parameter is needed to add a velocity axis to the output array model_values, see below:
-
RestFreq: rest frequency in MHz (default: 0). (If this parameter is set to zero, the intensity is plotted against frequency (in MHz) otherwise against velocity (in km s-1).
Output parameters:
-
input_file: the contents of the molfit file containing the parameters for the best fit.
-
model_values: the model function values for each data point, which correspond to the best fit. If RestFreq is unequal zero, the myXCLASSFit function adds a column to the output parameter model_values which contains the velocities. So, for a rest frequency unequal zero, the parameter model_values represents a python array with three columns, where the first column describes the frequencies, the second column describes the velocities and the third column the corresponding intensities.
-
JobDir: absolute path of the job directory created for the current run.
Example:
- fit observational data with a synthetic spectrum (uses MAGIX)
# In CASA: NumberIteration = 100 MolfitsFileName = 'my_molecules.molfit' experimentalData = 'my_observation.xml' AlgorithmXMLFile = 'my_algorithm-settings.xml' newmolfit, modeldata, JobDir = myXCLASSFit()
Output:
Start function myXCLASSFit: Creating job directory for current myXCLASS run: path-to-xclass/run/myXCLASSFit/job__14-06-2017__22-52-56__80078857/ .. done! All files of the current myXCLASSFit run are stored here! Analyze experimentalData parameter .. Analyze molfit parameter .. done! Analyze molfit file .. done! Check, if names of molecules in molfit file are included in myXCLASS database .. done! Creating io_control file .. done! Copy algorithm file to working directory .. done! Start MAGIX .. Read i/o control file. Open control file: path-to-xclass/run/myXCLASSFit/job__14-06-2017__22-52-56__80078857/io_control.xml Reading control parameters .. done! experimental_data: path-to-xclass/run/myXCLASSFit/job__14-06-2017__22-52-56__80078857/my_observation.xml parameter_file: path-to-xclass/run/myXCLASSFit/job__14-06-2017__22-52-56__80078857/parameters.xml fit_control: path-to-xclass/run/myXCLASSFit/job__14-06-2017__22-52-56__80078857/algorithm_control.xml fit_log: path-to-xclass/run/myXCLASSFit/job__14-06-2017__22-52-56__80078857/fit.log model_description: path-to-xclass/programs/MAGIX/Fit-Functions/myXCLASS/xml/myNewXCLASS.xml Import experimental file using settings declared in the xml-file. Open xml-file: path-to-xclass/run/myXCLASSFit/job__14-06-2017__22-52-56__80078857/my_observation.xml for experimental file import. Reading file .. Import settings: NumberExpFiles = 1 Experimental file number: 1 FileNamesExpFiles[0] = path-to-xclass/run/myXCLASSFit/job__14-06-2017__22-52-56__80078857/observed_spectrum.dat NumberExpRanges[0] = 1 MinExpRange[0][0] = 219764.24 MaxExpRange[0][0] = 221471.74 Reading ASCII-file path-to-xclass/run/myXCLASSFit/job__14-06-2017__22-52-56__80078857/observed_spectrum.dat .. done! File Nr.: 1 LengthExpRange[1] = 683 ExpDataX[1][0] = [ 219764.24] ExpDataX[1][LengthExpRange] = [ 221471.74] ExpDataY[1][0] = [ 0.1191] ExpDataY[1][LengthExpRange] = [ 0.0020452] Read the xml-file containing the start values for the fit process. Open xml-file: path-to-xclass/run/myXCLASSFit/job__14-06-2017__22-52-56__80078857/parameters.xml Reading file .. done! FitParametersNumber = 48 FitParameters = [[1, 0, 699892.916384, 0.0, 1, 1, 'CH3CN;v=0;', 1, 1, 'NEW', 'E', 'F', 'F', 'y', 0.1, 20.0, 3.0, 'y', 50.0, 550.0, 300.0, 'y', 14.0, 20.0, 15.3010299957, 'y', 4.0, 12.0, 8.0, 'y', -2.0, 2.0, -0.3, 'c', 2, 0, 'CH3CN;v8=1;', 'CH3CN;v=0;', 1, -1000000000.0, -1000000000.0, ' ', 'CH3C-13-N;v=0;', 'CH3CN;v=0;', 16, -1000000000.0, -1000000000.0, ' '], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], ['0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '0', '-1e+99', '-1e+99', '-1e+99', '0.1', '-1e+99', '-1e+99', '-1e+99', '50.0', '-1e+99', '-1e+99', '-1e+99', '14.0', '-1e+99', '-1e+99', '-1e+99', '4.0', '-1e+99', '-1e+99', '-1e+99', '-2.0', '-1e+99', '0', '0', '-1000000000.0', '-1000000000.0', '-1000000000.0', '-1000000000.0', '-1000000000.0', '-1000000000.0', '-1000000000.0', '-1000000000.0', '-1000000000.0', '-1000000000.0', '-1000000000.0', '-1000000000.0'], ['1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1e+99', '1e+99', '1e+99', '20.0', '1e+99', '1e+99', '1e+99', '550.0', '1e+99', '1e+99', '1e+99', '20.0', '1e+99', '1e+99', '1e+99', '12.0', '1e+99', '1e+99', '1e+99', '2.0', '1e+99', '10000', '10000', '1000000000.0', '1000000000.0', '1000000000.0', '1000000000.0', '1000000000.0', '1000000000.0', '1000000000.0', '1000000000.0', '1000000000.0', '1000000000.0', '1000000000.0', '1000000000.0']] FitParameterName = ['MinNumTransitionsSQL[[1]]', 'MaxNumTransitionsSQL[[1]]', 'MaxElowSQL[[1]]', 'MingASQL[[1]]', 'OrderTransSQL[[1]]', 'Number_Molecules[[1]]', 'Molecule_Name[[1]]', 'Molecule_Inex[[1]]', 'Number_MolLine[[1]]', 'MolfitFileFormatVersion[[1]]', 'EmissionFlag[[1]]', 'tdFlag[[1]]', 'nHFlag[[1]]', 'source_size_flag[[1]]', 'source_size_uplimit[[1]]', 'source_size_lowlimit[[1]]', 'source_size[[1]]', 'T_rot_flag[[1]]', 'T_rot_uplimit[[1]]', 'T_rot_lowlimit[[1]]', 'T_rot[[1]]', 'N_tot_flag[[1]]', 'N_tot_uplimit[[1]]', 'N_tot_lowlimit[[1]]', 'N_tot[[1]]', 'V_width_flag[[1]]', 'V_width_uplimit[[1]]', 'V_width_lowlimit[[1]]', 'V_width[[1]]', 'V_off_flag[[1]]', 'V_off_uplimit[[1]]', 'V_off_lowlimit[[1]]', 'V_off[[1]]', 'CFFlag[[1]]', 'NumberOfISOLines[[1]]', 'NumberOfGlobalISORatios[[1]]', 'IsoMolecule[[1]]', 'IsoMasters[[1]]', 'IsoRatio[[1]]', 'IsoLowerLimit[[1]]', 'IsoUpperLimit[[1]]', 'IsoGlobalParameters[[1]]', 'IsoMolecule[[2]]', 'IsoMasters[[2]]', 'IsoRatio[[2]]', 'IsoLowerLimit[[2]]', 'IsoUpperLimit[[2]]', 'IsoGlobalParameters[[2]]'] Start fit process. Open fit-control file: path-to-xclass/run/myXCLASSFit/job__14-06-2017__22-52-56__80078857/algorithm_control.xml Reading fit control parameters .. done! NumberOfFitAlgorithms: 1 algorithm: levenberg-marquardt Special algorithm settings: [2, 0.001] chilm: 1e-06 numrange: 0 numiter: 10 NumberProcessors: 6 MPI host file: DeterminationChi2: default PlotOption: ['normal', 'Rest Frequency [MHz]', 'T_mb [K]', 'z-axis', 'no'] Renormalized Chi^2: yes Read the xml-description of the input and the output file of the chosen fit-function module. Open xml-description stored in file: path-to-xclass/programs/MAGIX/Fit-Functions/myXCLASS/xml/myNewXCLASS.xml The maximum number of groups in the registration file is set to MaxNumberOfGroups = 100 If you use more groups in the registration file, please increase the variable MaxNumberOfGroups defined in line 2339 of the file Modules/magix-parts_python/FittingEngine.py Reading registration xml-file .. done! WARNING: The defined number of processors (6) is larger than the number of available processors (4). Reduce number of processors to the number of available processors. Number of Processors: 4 Writing registration mask for the fit model .. done! Using optimized version of MAGIX for myXCLASS (SMP): Initialize myXCLASS program: Reading parameter for myXCLASS from paramset variable .. done! Reading partition functions for all molecules from sqlite3 database .. done! Number of entries = 3 First Molecule in database: CH3CN;v=0; Last Molecule in database: CH3C-13-N;v=0; Reading parameters for radiative transitions for all molecules from sqlite database .. done! Number of transitions for each frequency range and molecule: Frequency range: 2.197642400000000E+05 MHz - 2.214717400000000E+05 MHz: Number of transitions for molecule "CH3CN;v=0;": 36 Number of transitions for molecule "CH3CN;v8=1;": 1053 Number of transitions for molecule "CH3C-13-N;v=0;": 46 Temporary files are stored in: /dev/shm/moeller/job_80079048/ Start Levenberg-Marquardt algorithm (SMP version) .. Renormalized limit for chi^2 = 6.1500000000E-04 Using Numerical Recepies (NR) version! Iteration: chi^2: alamda: Parameter: 1 2.165674686014673E+02 1.E-04 3.000000000000000E+00, 3.484876833700959E+02, 1.521629684579954E+01, 8.581881161608271E+00, -3.844639951123868E-01 2 2.165674686014673E+02 1.E-03 3.000000000000000E+00, 3.484876833700959E+02, 1.521629684579954E+01, 8.581881161608271E+00, -3.844639951123868E-01 3 8.819594167461759E+01 1.E-04 3.000000000000000E+00, 4.192060427256997E+02, 1.516249216535065E+01, 9.254406523524336E+00, -4.819527673659468E-01 4 8.819594167461759E+01 1.E-03 3.000000000000000E+00, 4.192060427256997E+02, 1.516249216535065E+01, 9.254406523524336E+00, -4.819527673659468E-01 5 5.002649280486062E+01 1.E-04 3.000000000000000E+00, 5.192922721910660E+02, 1.514887917909892E+01, 9.931117784803558E+00, -5.529279004974829E-01 6 5.002649280486062E+01 1.E-03 3.000000000000000E+00, 5.192922721910660E+02, 1.514887917909892E+01, 9.931117784803558E+00, -5.529279004974829E-01 7 5.002649280486062E+01 1.E-02 3.000000000000000E+00, 5.192922721910660E+02, 1.514887917909892E+01, 9.931117784803558E+00, -5.529279004974829E-01 8 4.932427271467029E+01 1.E-03 3.210336221785408E+00, 5.192922721910660E+02, 1.515274085964414E+01, 1.048288522241226E+01, -5.591081491134849E-01 9 4.932427271467029E+01 1.E-02 3.210336221785408E+00, 5.192922721910660E+02, 1.515274085964414E+01, 1.048288522241226E+01, -5.591081491134849E-01 10 4.932427271467029E+01 1.E-01 3.210336221785408E+00, 5.192922721910660E+02, 1.515274085964414E+01, 1.048288522241226E+01, -5.591081491134849E-01 Iteration stopped. Number of iterations is equal to max. number of iterations = 10 Sorting chi2-log file .. done! Finished Levenberg-Marquardt algorithm! Write values of the fit function to files. Writing data to ASCII file path-to-xclass/run/myXCLASSFit/job__14-06-2017__22-52-56__80078857/observed_spectrum.LM.out.dat .. done! Writing chi^2 values to file path-to-xclass/run/myXCLASSFit/job__14-06-2017__22-52-56__80078857/observed_spectrum.LM.out.chi2.dat .. done! Write optimized model parameter to file. Open xml-file: path-to-xclass/run/myXCLASSFit/job__14-06-2017__22-52-56__80078857/parameters.LM.out.xml Writing file .. done ! Program finished! Read in optimized input file(s) .. Read in model function values from file(s) .. Assuming, that the model function values for the best fit are stored to the variable "model_values", the output array "model_values" contains the model function values for the experimental data file observed_spectrum.LM.out.dat Please note, all files created by the current fit process (new molfit file, log-files etc.) are stored in the myXCLASSFit working directory "path-to-xclass/run/myXCLASSFit/job__14-06-2017__22-52-56__80078857/"!
Example scripts:
"my_fit_INcasa__xml.py", "my_fit_OUTcasa__xml..py"